Abstract
The ETS family of genes encode transcription factors (TFs) containing the ETS DNA binding domain, which have roles in cellular growth and development, including embryonic hematopoiesis. Dysregulation of these genes is associated with malignant transformation and tumorigenesis. In childhood acute myeloid leukemia (AML), the MNX1-ETV6 t(7;12)(q36;p13) and FUS-ERG t(16;21)(p11;q22) fusions are associated with adverse outcome. However, the biological and clinical implications of other ETS oncofusions in pediatric AML remain unknown.
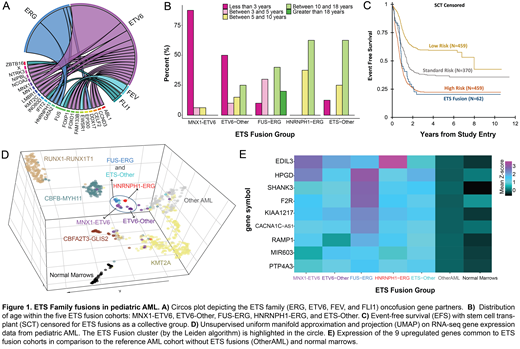
We identified 62 ETS gene fusions in 1,473 primary diagnostic AML samples (4.2%) using whole transcriptome RNA-sequencing and karyotype data. Four ETS family genes were identified to be involved in fusions with various partner genes: ETV6, ERG, FEV, and FLI1. Fusions with ETV6 were the most abundant (58%, 36/62) while FEV and FLI1 were the least; 21 fusion partners were identified where many function as TFs and co-activators (Figure 1A). MNX1-ETV6 (N=16) is enriched in infants (87.5% < 3 years, p < 0.001, Figure 1B), consistent with previous reports. ETV6 fusions with various partners (ETV6-Other, N=20) were likewise enriched in infants (50% < 3 years) and were significantly younger than patients without ETS fusions (median age: 3.0 vs 9.6 years, p = 0.025). FUS-ERG (N=10), HNRNPH1-ERG (N=8), and ETS fusions with various partners (ETS-Other, N=8) were primarily identified in patients 5-18 years old.
Outcomes for patients with ETS fusions was determined after censoring for stem cell transplantation. Patients with ETS fusions as a collective group had adverse outcome with an EFS of 17.7% vs 39.9% (p = 0.047, Figure 1C), similar to high-risk cases in the reference group (22%), suggesting that patients with any ETS fusion may be considered high risk. Evaluation of outcomes by individual fusion groups at 3 years from diagnosis demonstrated an EFS of 0% for HNRNPH1-ERG, 18.0% for FUS-ERG, 14.2% for ETV6-Other, 20.8% for ETS-Other, and 28.4% for those with MNX1-ETV6. Twenty-one patients (37.5%) with ETS family fusions received stem cell transplant (SCT) in first complete remission, including HNRNPH1-ERG (N=5), FUS-ERG (N=3), ETV6-Other (N=5), ETS-Other (N=2), and MNX1-ETV6 (N=6). For these SCT recipients, OS at 3 years after transplant was 60.2%.
We investigated whether ETS family fusions might have similar transcriptome profiles. Unsupervised uniform manifold approximation and projection (UMAP) on RNA-seq gene expression data followed by Leiden clustering found that individual fusions clustered in the same 3D space. More importantly, ETS fusion groups clustered closely to one another, indicating a shared transcriptional profile (Figure 1D, circle). Next, ETS fusion groups were each independently compared to the reference cohort (N=1421) using differential expression (DE) analysis. Intersection of DE genes revealed 17 overexpressed genes common to ETS fusions and 9/17 (52%) were also dysregulated when contrasting ETS cohorts to healthy normal marrows' transcriptome (N=68, Figure 1E). The minimal set of dysregulated genes included an adhesion molecule EDIL3, a prostaglandin (PG) enzyme HPGD, and a tyrosine phosphatase PTP4A3, which is strongly associated with progression in lymphoblastic leukemia and multiple myeloma. EDIL3 was reported to be overexpressed in MNX1-ETV6 and we found this molecule is a common feature of ETS fusions and their cellular dysregulation. The minimal set of 9 genes were further investigated using protein interaction networks defined from Pathway Commons v11. FUS-ERG and HNRNPH1-ERG both had significantly (adj. p < 0.001) activated HPGD networks; PG-E synthase and > 10 PG metabolism genes were upregulated. PG metabolism has important roles in regulating hematopoietic stem and progenitor (HSPC) functions and PG-E2 was shown to increase HSPC survival. ETV6-MXN1, ETV6-Other, and FUS-ERG had activated PTP4A3 networks and its expression was associated with sensitivity to BET inhibitors (BETi) in myeloma. They also exhibited increased activity of an ERG network with the overexpression of upstream regulators CBFA2T3 and GATA, and downstream targets like VWF and ZBTB16.
Overall, we show that ETS fusions are uniformly high risk and share dysregulated cell adhesion (EDIL3) and transcriptional networks for ERG, HPGD, and PTP4A3, which provide opportunities for further research into the metabolome and therapeutics (BETi) in these fusions.
Shaw: T-Cell and/or Gene Therapy for Cancer: Patents & Royalties.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal